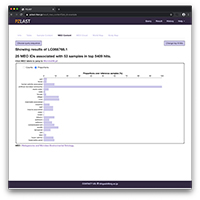
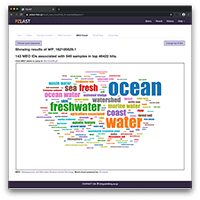
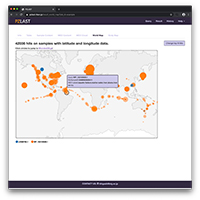
Using PZLAST, users can search the protein sequences to several Tera-bytes of public metagenomic amino acid sequences in approximately 10-20 minutes.
PZLAST is built on a ZettaScaler-3.0 Server Unit [1][2] with PEZY-SC3 Processor [3] [4]. The algorithm of PZLAST is basically similar to CLAST [5].
References
ZettaScaler-3.0 and PEZY-SC3
[1] PZLAST Meta genome analysis system
[2] ZettaScaler-3.0
[3] PEZY-SC3
[4] Naoya Hatta, Shuntaro Tsunoda, Kouhei Uchida, Taichi Ishitani, Ryota Shioya, Kei Ishii. PEZY-SC3: A MIMD Many-core Processor for Energy-efficient Computing. https://doi.org/10.48550/arXiv.2301.07510
CLAST
[5] CLAST: CUDA implemented large-scale alignment search tool
Citation
If you find PZLAST useful in your research, please cite the following papers:
Hiroshi Mori, Hitoshi Ishikawa, Koichi Higashi, Yoshiaki Kato, Toshikazu Ebisuzaki, Ken Kurokawa, PZLAST: an ultra-fast amino acid sequence similarity search server against public metagenomes, Bioinformatics, 2021;, btab492, https://doi.org/10.1093/bioinformatics/btab492
Hitoshi Ishikawa, Hiroshi Mori, Koichi Higashi, Yoshiaki Kato, Tomofumi Sakai, Toshikazu Ebisuzaki, Ken Kurokawa. PZLAST: an ultra-fast sequence similarity search tool implemented on a MIMD processor. Special Issue on the Ninth International Symposium on Networking and Computing, 2022 Volume 12 Issue 2 Pages 446-466. https://doi.org/10.15803/ijnc.12.2_446
- Contact
- This web site is operated by Genome Evolution Laboratory at the National Institute of Genetics.
- If you have any question, request, or suggestion, please contact the E-mail address below.
- khigashinig.ac.jp
- v. 0.4.0 (2024-01-20)
- The computation nodes have been revamped and the computation speed is even faster than in previous versions.
- v. 0.3.1 (2022-07-20)
- Added "Sample-wise results" to the results page.
- Completion-table, -MEO cloud, -world map results are organized into the "Sample-wise results" tab.
- v. 0.3.0 (2022-07-13)
- Added two features to the results page.
- One is environmental coverage information for each amino acid residue, and the other is a table of query hit fulfillment rates for each sample.
- v. 0.2.1 (2021-12-17)
- Reference database has been updated.
- As the number of reference sequences increased from 42 Billion to 64 Billion, the processing time of the search increased by about 20%.
- v. 0.2.0 (2021-8-20)
- Alignment information has been added to the output table.
- v. 0.1.0 (2020-4-1)
- PZLAST Web Service: First release.